
Two Bates College scientists have received nearly $373,000 from the National Institutes of Health for first-of-its-kind research into the genetic functioning of bacteria that cause Lyme disease.
The project draws on new capabilities in high-resolution microscopy that make it possible to observe spatial arrangements of RNA in Borrelia burgdorferi, one of four Borrelia species that cause Lyme disease. This study of variations in RNA location and shape could eventually suggest new medical responses to Lyme, which strikes up to 300,000 people in the U.S. annually.
Receiving the $372,639 grant for the two-year project are physicist Travis Gould, an expert in the field of fluorescence nanoscopy, and biochemist Paula Schlax, who researches gene expression in spiral-shaped bacteria, also known as spirochetes, such as Lyme bacteria.
Biochemist Paula Schlax and physicist Travis Gould pose with Gould’s STED super-resolution microscope in Carnegie Science Hall. (Phyllis Graber Jensen/Bates College)
“RNA is an intermediate in the process of cells making proteins,” says Schlax, a professor of chemistry and biochemistry at Bates. “We’re trying to understand generally how production of proteins gets turned on and off when the bacteria move from ticks to mammals and from mammals back to ticks” — changes in the bacteria’s environment that change the shape and location of RNA.
“We know from other bacteria that RNA’s location inside the cell seems to affect how long that RNA lasts — whether it’s near the edges of the cell, or the ends of the cell, or spread out evenly inside. Our hypothesis is that how fast RNA gets broken down, or doesn’t get broken down, probably helps the cell decide which proteins to make when conditions change, such as when the bacteria moves from the tick to a mammal or vice versa.”
Variations in protein production could cause variations in the bacteria’s disease-causing capability. “The more we understand that process,” Schlax says, “the easier it is to think about new targets for drugs and new therapeutics.”
Until quite recently, the physical limitations of microscope technology curtailed its usefulness in testing such a hypothesis. The bacteria have a characteristic shape, says Gould: very skinny in relation to length. The length is typically around 20 microns, or millionths of a meter, but the bacteria’s internal diameter is vastly smaller, at about 200 nanometers, or billionths of a meter. (A piece of paper is about 100,000 nanometers thick.)
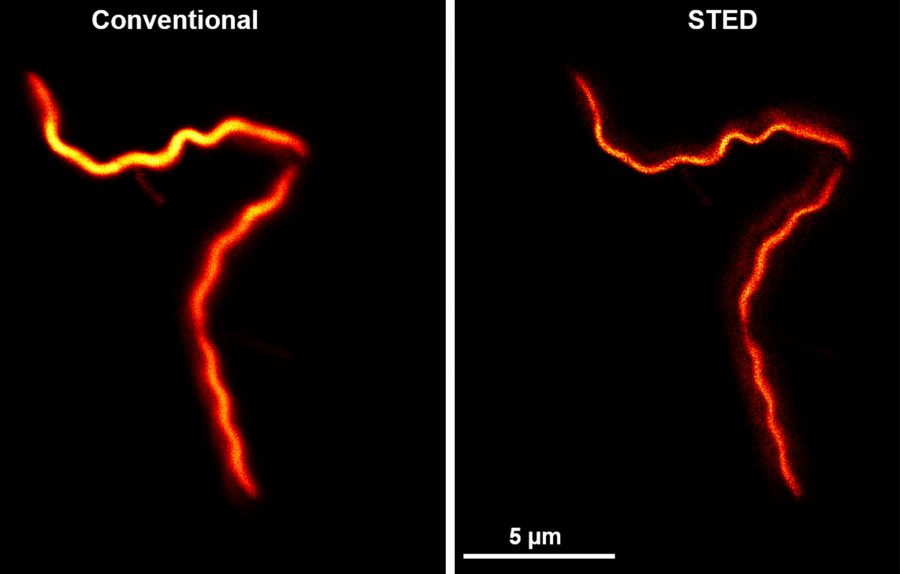
“That 200 nanometers is, in the best-case scenario, at the limit of a conventional microscope’s resolution,” Gould says. “So a conventional microscope can’t answer these questions about where RNA is within that 200-nanometer cylinder.”
But Gould, an associate professor of physics, is an innovator in imaging technologies that use lasers, fluorescing molecules, and other means to attain much higher resolution. For the NIH-funded research, he has adapted an existing Bates microscope that he built and that uses a process called stimulated emission depletion, or STED, to capture images of the B. burgdorferi RNA.
Going from older microscopy technologies to Gould’s newly updated STED “is like putting on glasses for the first time.”
Specifically, he added another laser to the instrument that expands its imaging capability from two to three dimensions. Going from older technologies to this latest iteration, says Schlax, “is like putting on glasses for the first time.”
Complementary to the STED technology, the researchers and their students will use a technique called fluorescence in situ hybridization (FISH) that deploys fluorescent probes to specific parts of the “transcripts” that the microbe’s DNA imparts to its RNA.
The research will be the first to identify patterns of transcript localization within B. burgdorferi, and, notably, the first research to use STED microscopy for this sort of localization within any spirochete.

This is significant given the range and impact of diseases caused by such bacteria, including syphilis, yaws, periodontal disease, and leptospirosis, whose effects include kidney failure.
Joining Schlax and Gould in the project are Bates students and research associate Anna Bowsher, whose position is funded by the NIH grant. The work entails growing B. burgdorferi microbes in the lab, affixing individual cells to slides, and introducing DNA molecules, complete with fluorescent tags, that are tailored to activate a specific RNA response.
Then the slides will be examined with Gould’s STED microscope, and the results compiled into a spatial-distribution analysis of different types of RNAs. The team hopes that they will have results to report by summer 2020.

The project will involve both thesis students advised by Gould and Schlax and students doing summer research. “These kinds of projects really are great for students to see how science is done,” says Schlax, “and hopefully get their names on some papers and keep them interested in science.”
With the use of fluorescing molecules now standard practice in high-resolution microscopy, STED imaging achieves enhanced resolution through a technique of selectively switching off such molecules. STED is one of a number of so-called super-resolution techniques developed to bypass the diffraction limit, a limit on the resolution of conventional microscopy imposed by the length of light waves.
Gould estimates that all told, there are likely two dozen or so labs equipped with commercially available STED microscopes, and another handful that use custom-built instruments like his.




