Research
Bates College is an undergraduate liberal arts college, and as such, the majority of the research in my lab is conducted by students. They are engaged in every facet from hypothesis generation, data collection, data analysis, and manuscript writing. Please feel free to contact me to learn about the exciting opportunities for research during the school year and in the summer.
Projects in the Lab
Role of NF-E2-related factor (Nrf) proteins in the developmental process
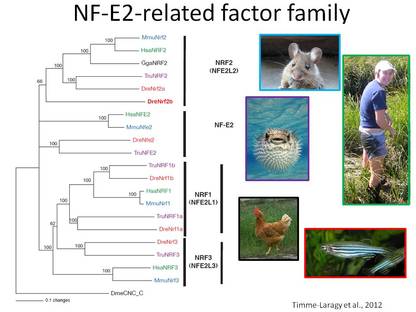 Transcription factors in the CNC-bZIP family, NFE2, NRF1, NRF2 and NRF3, regulate genes with a wide range of functions in response to both physiological and exogenous signals, including those indicating changes in cellular redox status. Given their role in helping to maintain cellular homeostasis, it is imperative to understand the expression, regulation, and function of CNC-bZIP genes during embryonic development. We are exploring the expression and function of four of the six nrf genes (nfe2, nrf1a, nrf1b, and nrf3) using zebrafish embryos as a model system. Our collaborators are at Amherst College and UMass Amherst.
Transcription factors in the CNC-bZIP family, NFE2, NRF1, NRF2 and NRF3, regulate genes with a wide range of functions in response to both physiological and exogenous signals, including those indicating changes in cellular redox status. Given their role in helping to maintain cellular homeostasis, it is imperative to understand the expression, regulation, and function of CNC-bZIP genes during embryonic development. We are exploring the expression and function of four of the six nrf genes (nfe2, nrf1a, nrf1b, and nrf3) using zebrafish embryos as a model system. Our collaborators are at Amherst College and UMass Amherst.
Transcription factor cross-talk in developmental toxicity
 Sensitivity to toxicants is often enhanced during embryonic development, when exposure to low levels of chemicals can disrupt developmental process and lead to disease. Chemically-activated transcription factors act as environmental biosensors of external toxic stimuli and regulate gene expression involved in mitigating negative effects. Rarely is the response regulated by one transcription factor; rather, it is generally a coordinated effort between two or more transcription factors. One such interaction that may play an important role in the embryonic response to toxicant exposure is between two ligand-activated transcription factors — the aryl hydrocarbon receptor (AHR) and nuclear factor erythroid 2-related factor 2 (Nrf2). Ahr and Nrf2 independently regulate many genes involved in the cellular response to toxicants and oxidative stress as well as development, apoptosis, and differentiation. Because Ahr and Nrf2 regulate diverse gene targets, the significance of such crosstalk during development could be large; however, their crosstalk has only been characterized in adult mammals and cell culture. Zebrafish are an excellent model to study these interactions during development because of their rapid and external development as transparent embryos, the availability of extensive genomic and molecular tools, and conservation of developmental signaling pathways with humans, which can facilitate the extrapolation of results. This research uses a collaborative and multidisciplinary approach to elucidate Ahr-nrf2 crosstalk and, in particular, the role of Ahr in regulating transcription of nrfs and their downstream genes during development and in response to toxicant exposure.
Sensitivity to toxicants is often enhanced during embryonic development, when exposure to low levels of chemicals can disrupt developmental process and lead to disease. Chemically-activated transcription factors act as environmental biosensors of external toxic stimuli and regulate gene expression involved in mitigating negative effects. Rarely is the response regulated by one transcription factor; rather, it is generally a coordinated effort between two or more transcription factors. One such interaction that may play an important role in the embryonic response to toxicant exposure is between two ligand-activated transcription factors — the aryl hydrocarbon receptor (AHR) and nuclear factor erythroid 2-related factor 2 (Nrf2). Ahr and Nrf2 independently regulate many genes involved in the cellular response to toxicants and oxidative stress as well as development, apoptosis, and differentiation. Because Ahr and Nrf2 regulate diverse gene targets, the significance of such crosstalk during development could be large; however, their crosstalk has only been characterized in adult mammals and cell culture. Zebrafish are an excellent model to study these interactions during development because of their rapid and external development as transparent embryos, the availability of extensive genomic and molecular tools, and conservation of developmental signaling pathways with humans, which can facilitate the extrapolation of results. This research uses a collaborative and multidisciplinary approach to elucidate Ahr-nrf2 crosstalk and, in particular, the role of Ahr in regulating transcription of nrfs and their downstream genes during development and in response to toxicant exposure.
Population Genetics of Invasive Crabs in the Gulf of Maine
 The European green crab, Carcinus maenas, is one of the most successful invasive marine species. Since its arrival in New York 200 years ago, it has spread North along the coast, all the way to Nova Scotia, establishing populations and affecting local communities. Recent invasions to Nova Scotia have brought in a new, more temperature tolerant and possibly more voracious, strain of C. maenas. The Gulf of Maine has seen a recent increase in C. maenas, which have been causing negative affects on local communities and the shellfish industry. In order to asses the population structure of C. maenas in the Gulf of Maine, our studies use polymorphisms in the cytochrome oxidase I (COI) gene to see whether the northern strain has spread South and, thus, is potentially responsible for these affects. We are now using these same techniques to study the invasion of the Asian Shore Crab, Hemigrapsus sanguineus, along the East Coast of the US.
The European green crab, Carcinus maenas, is one of the most successful invasive marine species. Since its arrival in New York 200 years ago, it has spread North along the coast, all the way to Nova Scotia, establishing populations and affecting local communities. Recent invasions to Nova Scotia have brought in a new, more temperature tolerant and possibly more voracious, strain of C. maenas. The Gulf of Maine has seen a recent increase in C. maenas, which have been causing negative affects on local communities and the shellfish industry. In order to asses the population structure of C. maenas in the Gulf of Maine, our studies use polymorphisms in the cytochrome oxidase I (COI) gene to see whether the northern strain has spread South and, thus, is potentially responsible for these affects. We are now using these same techniques to study the invasion of the Asian Shore Crab, Hemigrapsus sanguineus, along the East Coast of the US.
Cause of eelgrass decline around Mount Desert Island, Maine
 Along with Jane Disney of The MDI Biological Laboratory, I have been investigating the cause of eelgrass decline around Mount Desert Island. Zostera marina or “eelgrass” is a common seagrass found widespread in northern coastlines across the world. Eelgrass beds serve a vital role in marine ecosystems of Maine, protecting shorelines against erosive forces and providing an ecosystem for larval and juvenile fish and shellfish. Between 1996 and 2013, the coverage of eelgrass in the upper Frenchman Bay area of Mount Desert Island, ME, was reduced from 3,174 to between 75 and 183 acres. Contrasting sites with healthy populations to sites that have sustained significant loss is key to identifying decline causation. To date, we have investigated the role of eelgrass genetic diversity as well as green crab predation in the decline.
Along with Jane Disney of The MDI Biological Laboratory, I have been investigating the cause of eelgrass decline around Mount Desert Island. Zostera marina or “eelgrass” is a common seagrass found widespread in northern coastlines across the world. Eelgrass beds serve a vital role in marine ecosystems of Maine, protecting shorelines against erosive forces and providing an ecosystem for larval and juvenile fish and shellfish. Between 1996 and 2013, the coverage of eelgrass in the upper Frenchman Bay area of Mount Desert Island, ME, was reduced from 3,174 to between 75 and 183 acres. Contrasting sites with healthy populations to sites that have sustained significant loss is key to identifying decline causation. To date, we have investigated the role of eelgrass genetic diversity as well as green crab predation in the decline.
Adaptation in natural populations to chronic pollution
 I am interested in how populations have adapted through natural selection to chronic pollution in their environment. During graduate school in Margie Oleksiak’s lab, I studied how populations of the estuarine killifish, Fundulus heteroclitus, have adapted to living in heavily polluted Superfund sites. In my lab at Bates, I am focusing on how the keystone plant species, the smooth cordgrass (Spartina alterniflora), has adapted to the same polluted environments.
I am interested in how populations have adapted through natural selection to chronic pollution in their environment. During graduate school in Margie Oleksiak’s lab, I studied how populations of the estuarine killifish, Fundulus heteroclitus, have adapted to living in heavily polluted Superfund sites. In my lab at Bates, I am focusing on how the keystone plant species, the smooth cordgrass (Spartina alterniflora), has adapted to the same polluted environments.
In order to detect selection in polluted populations, I have used an experimental design termed a “triad design”, where one polluted population is compared with two clean flanking populations. This experimental design distinguishes pollutant effects from demographic ones because the genetic distance between the two clean reference populations is greater than the genetic distance between the polluted population and either reference population. Thus, the variation due to demography is accounted for by comparing the polluted population to the combined variation in the northern and southern reference populations. Significant divergence in a polluted population compared to both paired reference populations suggests that pollution or other environmental factors associated with the Superfund sites are affecting the change in allele frequency of a particular genetic marker.